Hartline Lab Research - Details
Physiologically-accurate computer modeling
This area of research has progressed over time from fairly simple models of neurons
and network interactions to increasingly more complex and realistic ones:
- Early work involved simulation of simplified circuits of both pyloric and gastric
subnetworks in the stomatogastric ganglion (STG).
See Warshaw and Hartline (1976)
for details.
- Physiological work on
repetitive firing characteristics, time courses and strengths
of specific synaptic intereactions, and involvement of active membrane properties, led to
a more refined model for the pyloric subsystem. See
Hartline (1979)
- The previous model still lacked accuracy in several aspects of the coordinated firing
patterns. Many of these were traced to the absence of plateau properties in follower neurons.
Once plateaus had been characterized (see
Russell and Hartline (1982) and
Russell and Hartline (1984) the ground was laid for a more refined model. See
Hartline (1987)
- Modeling work since the last listed has focused on functions of individual cells (see next)
Computational characteristics of neuritic trees
(Collaboration with Dr. Katherine Graubard, Dept. of Zoology,
University of Washington)
Synaptic inputs to one part of a typically multiply-branched neuritic tree are subject to several
types of modification as they spread to other parts of the cell. The modifications they undergo
potentially differ significantly among various synaptic output sites and the spike-initiating regions
("trigger zones") of the cell. This research focus attempts to understand what sorts of signal
transformations occur in real neurons by applying modeling techniques to reconstructed STG cells.
The approach is briefly as follows:
- 1. Fluorescent dye injection
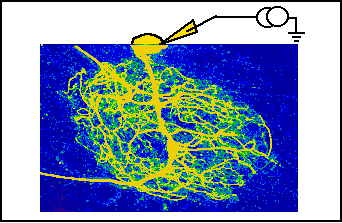
A fluorescent dye is injected into an identified neuron with a microelectrode
- 2. Confocal microscope imaging
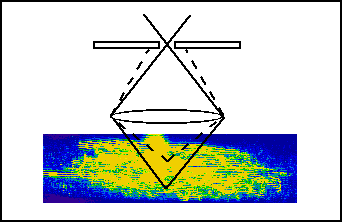
The ganglion is fixed histologically and scanned under a confocal microscope.
The result is a stack of images representing serial optical cross sections of the cell.
- 3. 3D Computer reconstruction
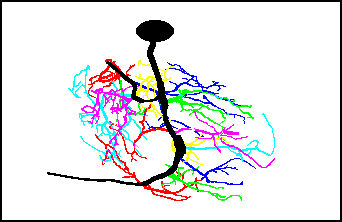
The fluorescent processes of the neurons are followed through the stack of optical
cross-sections using tree-tracing macros
written for NIH Image software (by Wayne Rasband) on a Macintosh computer. The software
records lengths, diameters, coordinates of segment ends, and segment connectivities to form
a 3D reconstruction of the neuron.
- 4. Computer modeling
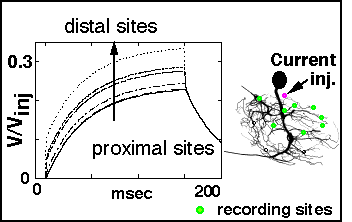
Data on segment dimensions and connectivities are fed to a program that converts the data
to a format compatible with the NEURON modeling program (by Michael Hines). A variety of
inputs in the form of current pulses or simulated synaptic activations can be placed in
various parts of the cell and the results of these perturbations can be followed at any
other desired point in the same cell.
- For a poster presenting some of the results of this study,
click here.
Space clamp errors in extended neurons
This project is developing approaches for assessing and correcting errors in voltage clamp
experiments that arise from a lack of perfect space clamp.
- [Detailed overview to appear here ... have patience!]
- For a poster presenting some of the results of this study,
click here.
 Return to Hartline Home Page.
Return to Hartline Home Page.
 Return to PBRC Home Page.
Return to PBRC Home Page.



